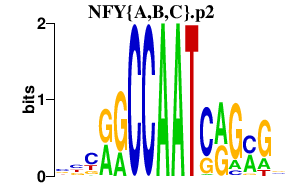
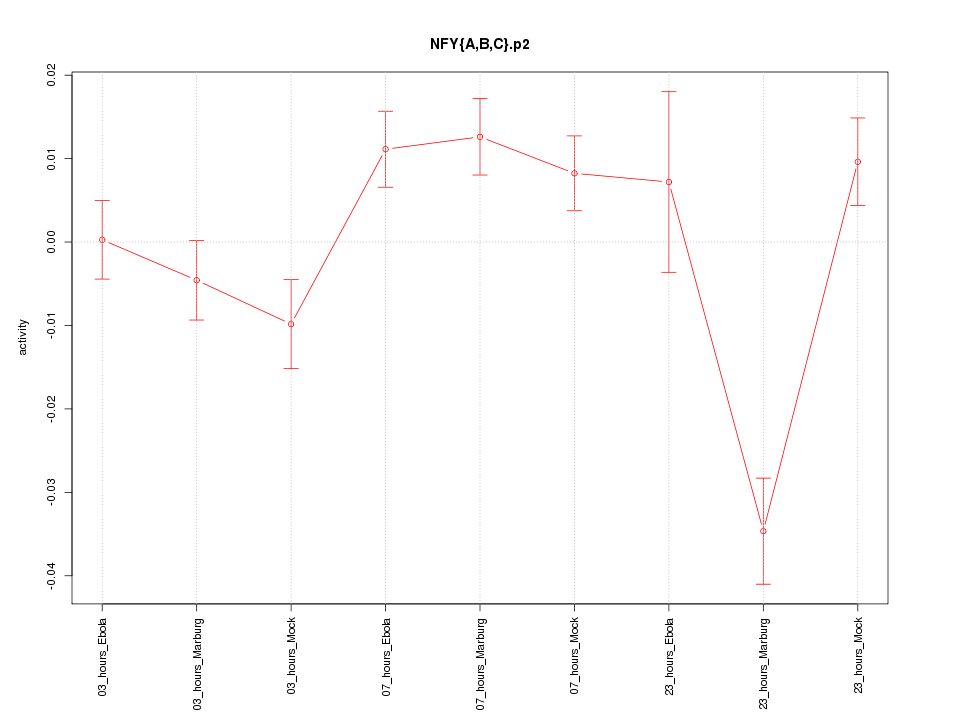
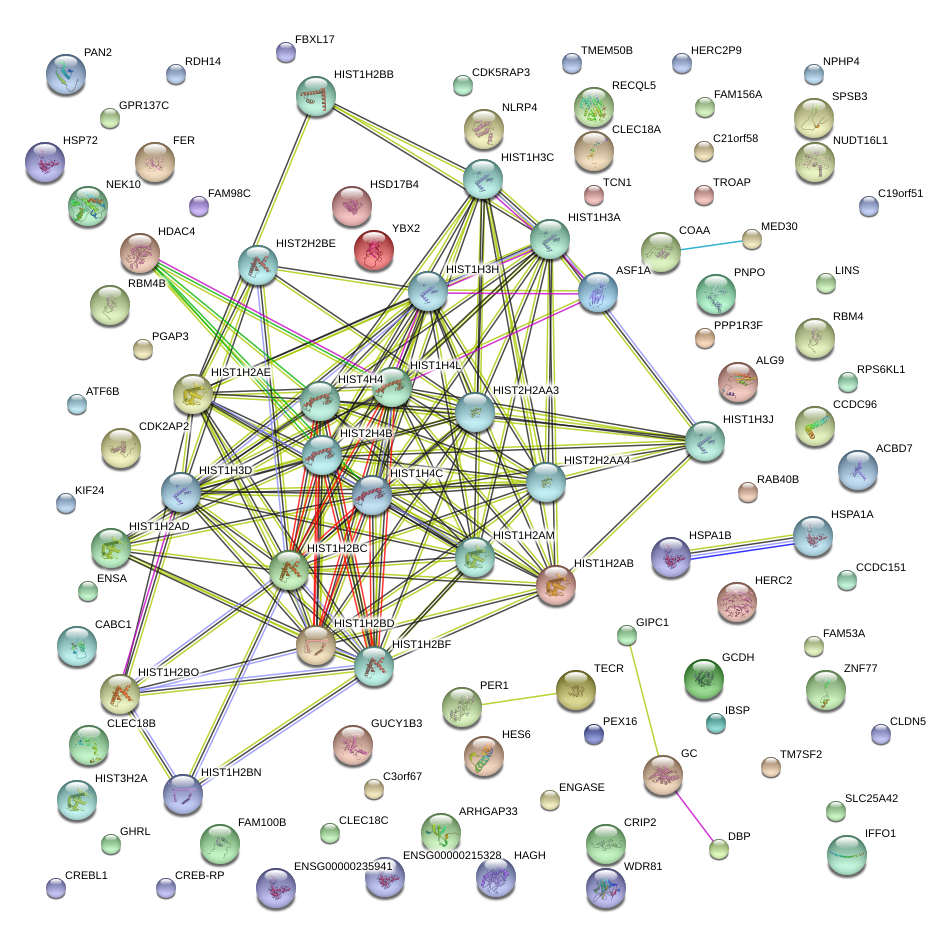
|
chr6_+_26045611
|
3.560
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr19_+_38893766
|
2.188
|
NM_174905
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr1_+_151763174
|
1.767
|
|
|
|
|
chr6_+_26199712
|
1.728
|
NM_003522
|
HIST1H2BF
|
histone cluster 1, H2bf
|
|
chr6_-_26043884
|
1.576
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr2_-_240322692
|
1.180
|
|
HDAC4
|
histone deacetylase 4
|
|
chr2_-_240322620
|
1.168
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr4_+_88720701
|
1.109
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr16_+_72698996
|
1.076
|
|
|
|
|
chr6_+_31783314
|
1.025
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr1_-_6052481
|
1.018
|
NM_015102
|
NPHP4
|
nephronophthisis 4
|
|
chr11_-_45939361
|
1.018
|
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chrX_+_49126299
|
1.010
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr17_-_8059637
|
1.009
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr10_-_15130773
|
1.007
|
NM_001039844
|
ACBD7
|
acyl-CoA binding domain containing 7
|
|
chr19_-_49140569
|
0.998
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr4_-_120375770
|
0.979
|
|
|
|
|
chr6_+_31783280
|
0.966
|
NM_005345
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr9_-_34329151
|
0.963
|
NM_194313
|
KIF24
|
kinesin family member 24
|
|
chr11_-_67275549
|
0.953
|
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr5_+_108084638
|
0.930
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr6_+_31783530
|
0.928
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chrX_+_49126326
|
0.910
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr21_-_34852282
|
0.902
|
NM_006134
|
TMEM50B
|
transmembrane protein 50B
|
|
chr19_+_38893804
|
0.902
|
|
FAM98C
|
family with sequence similarity 98, member C
|
|
chr17_+_74261280
|
0.901
|
NM_182565
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr19_+_19174792
|
0.893
|
NM_178526
|
SLC25A42
|
solute carrier family 25, member 42
|
|
chr17_-_80656461
|
0.884
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr21_-_34852226
|
0.877
|
|
TMEM50B
|
transmembrane protein 50B
|
|
chrX_+_49047887
|
0.872
|
|
|
|
|
chr6_+_31795395
|
0.871
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr16_-_1832576
|
0.854
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr16_+_4743693
|
0.829
|
NM_001193452
NM_032349
|
NUDT16L1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1
|
|
chr17_-_7197896
|
0.820
|
|
YBX2
|
Y box binding protein 2
|
|
chrX_+_49126480
|
0.816
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr19_-_2944894
|
0.803
|
NM_021217
|
ZNF77
|
zinc finger protein 77
|
|
chr17_+_77071002
|
0.801
|
NM_001042573
|
ENGASE
|
endo-beta-N-acetylglucosaminidase
|
|
chr3_-_27410852
|
0.799
|
NM_199347
|
NEK10
|
NIMA (never in mitosis gene a)- related kinase 10
|
|
chr16_-_1832571
|
0.785
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr19_-_14606939
|
0.776
|
NM_005716
NM_202467
NM_202468
NM_202469
NM_202470
NM_202494
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr16_-_74455239
|
0.754
|
NM_001011880
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chrX_+_52926334
|
0.741
|
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr16_+_4743709
|
0.733
|
|
NUDT16L1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1
|
|
chr19_+_36266366
|
0.730
|
NM_001172630
NM_052948
|
ARHGAP33
|
Rho GTPase activating protein 33
|
|
chr14_-_75389901
|
0.728
|
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1
|
|
chr19_-_14606893
|
0.721
|
|
GIPC1
|
GIPC PDZ domain containing family, member 1
|
|
chr1_-_6052410
|
0.721
|
|
NPHP4
|
nephronophthisis 4
|
|
chr16_-_1832304
|
0.706
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr1_+_227127937
|
0.698
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr19_+_14640413
|
0.697
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr1_-_149814264
|
0.695
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr21_-_47743753
|
0.686
|
NM_058180
|
C21orf58
|
chromosome 21 open reading frame 58
|
|
chr21_-_34852163
|
0.675
|
|
TMEM50B
|
transmembrane protein 50B
|
|
chr1_-_16161279
|
0.671
|
|
|
|
|
chr17_+_46048406
|
0.670
|
NM_176096
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr14_+_53019865
|
0.665
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr2_-_239148626
|
0.664
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr6_-_27858569
|
0.658
|
NM_003535
|
HIST1H3J
|
histone cluster 1, H3j
|
|
chr1_+_227127999
|
0.651
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr17_+_1619816
|
0.650
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr19_+_13001966
|
0.650
|
NM_000159
NM_013976
|
GCDH
|
glutaryl-CoA dehydrogenase
|
|
chr4_-_7044664
|
0.650
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chr12_-_56727720
|
0.639
|
NM_001166279
NM_014871
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr4_+_156680125
|
0.635
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr11_+_64879294
|
0.633
|
NM_003273
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr16_+_1833003
|
0.633
|
|
|
|
|
chr1_+_227127959
|
0.629
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr12_+_49716970
|
0.628
|
NM_001100620
NM_005480
|
TROAP
|
trophinin associated protein (tastin)
|
|
chr19_+_14640363
|
0.611
|
NM_138501
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr6_-_26033795
|
0.611
|
NM_003513
|
HIST1H2AB
|
histone cluster 1, H2ab
|
|
chr7_-_65235688
|
0.605
|
|
LOC441242
|
uncharacterized LOC441242
|
|
chr11_+_66406189
|
0.601
|
|
RBM4
|
RNA binding motif protein 4
|
|
chr12_-_6665220
|
0.596
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr19_+_14640406
|
0.595
|
|
TECR
|
trans-2,3-enoyl-CoA reductase
|
|
chr3_-_59035700
|
0.594
|
NM_198463
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr16_-_1876528
|
0.593
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr19_-_55677919
|
0.593
|
NM_178837
|
C19orf51
|
chromosome 19 open reading frame 51
|
|
chr8_+_118532953
|
0.587
|
NM_080651
|
MED30
|
mediator complex subunit 30
|
|
chr17_+_74261431
|
0.584
|
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr6_-_32095974
|
0.579
|
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr1_-_150602020
|
0.579
|
|
ENSA
|
endosulfine alpha
|
|
chr6_+_26104103
|
0.578
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chr5_-_107717798
|
0.577
|
NM_001163315
|
FBXL17
|
F-box and leucine-rich repeat protein 17
|
|
chr6_-_26197477
|
0.577
|
|
HIST1H3D
|
histone cluster 1, H3d
|
|
chr11_-_45939568
|
0.575
|
NM_004813
NM_057174
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr22_-_19511777
|
0.573
|
|
CLDN5
|
claudin 5
|
|
chr15_-_101142375
|
0.573
|
NM_001040616
|
LINS
|
lines homolog (Drosophila)
|
|
chr17_+_46019022
|
0.572
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chrX_+_52926358
|
0.569
|
|
FAM156B
|
family with sequence similarity 156, member B
|
|
chr14_+_105941061
|
0.567
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr7_-_65215903
|
0.564
|
|
|
|
|
chr4_-_1685717
|
0.564
|
NM_001013622
NM_001174070
|
FAM53A
|
family with sequence similarity 53, member A
|
|
chr15_-_20711386
|
0.564
|
|
HERC2P3
|
hect domain and RLD 2 pseudogene 3
|
|
chr6_-_32095993
|
0.563
|
NM_001136153
NM_004381
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr2_-_240323332
|
0.563
|
|
HDAC4
|
histone deacetylase 4
|
|
chr10_+_96522457
|
0.560
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr4_-_1722776
|
0.559
|
|
TMEM129
|
transmembrane protein 129
|
|
chr17_-_8055684
|
0.554
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr7_+_148982371
|
0.552
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr17_-_43025027
|
0.549
|
NM_001080443
|
KIF18B
|
kinesin family member 18B
|
|
chr19_-_10491172
|
0.549
|
|
TYK2
|
tyrosine kinase 2
|
|
chr1_+_145438437
|
0.547
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr11_-_568419
|
0.544
|
|
MIR210HG
|
MIR210 host gene (non-protein coding)
|
|
chr12_-_56727446
|
0.543
|
NM_001127460
|
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr1_-_36915969
|
0.543
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr11_-_47198379
|
0.541
|
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr20_+_13976263
|
0.534
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr3_-_120461350
|
0.531
|
|
|
|
|
chr1_+_192778168
|
0.530
|
NM_002923
|
RGS2
|
regulator of G-protein signaling 2, 24kDa
|
|
chr15_-_23378167
|
0.528
|
|
HERC2P2
|
hect domain and RLD 2 pseudogene 2
|
|
chr16_-_1661969
|
0.527
|
NM_014714
|
IFT140
|
intraflagellar transport 140 homolog (Chlamydomonas)
|
|
chr1_+_227127993
|
0.524
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr19_-_15529738
|
0.523
|
|
AKAP8L
|
A kinase (PRKA) anchor protein 8-like
|
|
chr17_-_8198605
|
0.520
|
|
SLC25A35
|
solute carrier family 25, member 35
|
|
chr6_-_27835308
|
0.518
|
NM_005322
|
HIST1H1B
|
histone cluster 1, H1b
|
|
chr22_+_42486904
|
0.515
|
|
LOC100132273
|
uncharacterized LOC100132273
|
|
chr22_-_37172113
|
0.515
|
NM_001177701
NM_006860
|
IFT27
|
intraflagellar transport 27 homolog (Chlamydomonas)
|
|
chr19_+_4969123
|
0.512
|
NM_015015
|
KDM4B
|
lysine (K)-specific demethylase 4B
|
|
chr3_-_186288331
|
0.511
|
NM_001134415
|
TBCCD1
|
TBCC domain containing 1
|
|
chr9_+_131798899
|
0.509
|
|
FAM73B
|
family with sequence similarity 73, member B
|
|
chr1_+_12079298
|
0.504
|
NM_021933
|
MIIP
|
migration and invasion inhibitory protein
|
|
chr6_-_41863086
|
0.502
|
NM_018561
|
USP49
|
ubiquitin specific peptidase 49
|
|
chr17_+_46048477
|
0.502
|
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr9_+_131843378
|
0.499
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr12_-_15091441
|
0.498
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr1_-_28241233
|
0.494
|
NM_002946
|
RPA2
|
replication protein A2, 32kDa
|
|
chr1_-_149859465
|
0.493
|
NM_175065
|
HIST2H2AB
HIST2H2BE
|
histone cluster 2, H2ab
histone cluster 2, H2be
|
|
chr11_-_47198420
|
0.491
|
|
ARFGAP2
|
ADP-ribosylation factor GTPase activating protein 2
|
|
chr17_-_58212896
|
0.490
|
|
|
|
|
chrX_-_52987610
|
0.488
|
NM_001242491
|
FAM156A
FAM156B
|
family with sequence similarity 156, member A
family with sequence similarity 156, member B
|
|
chr3_+_50229022
|
0.488
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr2_+_73144603
|
0.487
|
NM_004097
|
EMX1
|
empty spiracles homeobox 1
|
|
chr19_-_14629227
|
0.485
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr6_+_33172418
|
0.480
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr17_-_7307413
|
0.480
|
NM_152766
|
C17orf61-PLSCR3
C17orf61
|
C17orf61-PLSCR3 readthrough
chromosome 17 open reading frame 61
|
|
chr17_+_48172508
|
0.478
|
NM_001199900
NM_002611
NM_001199899
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr19_+_50031232
|
0.476
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr17_-_7197866
|
0.476
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr1_+_12079522
|
0.473
|
|
MIIP
|
migration and invasion inhibitory protein
|
|
chr3_+_111393522
|
0.473
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr16_+_57769667
|
0.472
|
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr1_+_16062808
|
0.466
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr17_-_7197811
|
0.465
|
|
YBX2
|
Y box binding protein 2
|
|
chr10_-_102790877
|
0.463
|
NM_001195263
NM_024895
|
PDZD7
|
PDZ domain containing 7
|
|
chr19_+_49622480
|
0.462
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr16_+_103892
|
0.461
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr2_-_178973065
|
0.458
|
NM_001077197
|
PDE11A
|
phosphodiesterase 11A
|
|
chr17_-_3539384
|
0.456
|
NM_013276
|
SHPK
|
sedoheptulokinase
|
|
chr7_-_72298752
|
0.456
|
NM_001145440
NM_001145441
|
TYW1B
|
tRNA-yW synthesizing protein 1 homolog B (S. cerevisiae)
|
|
chr4_+_4861371
|
0.456
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr5_-_64858931
|
0.452
|
NM_022145
|
CENPK
|
centromere protein K
|
|
chr6_-_25726754
|
0.452
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr1_+_64239566
|
0.452
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr17_+_46048467
|
0.451
|
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr11_+_66406087
|
0.451
|
NM_001198843
NM_001198844
NM_002896
|
RBM4
|
RNA binding motif protein 4
|
|
chr16_-_4852571
|
0.450
|
NM_024589
|
ROGDI
|
rogdi homolog (Drosophila)
|
|
chr1_-_26633110
|
0.450
|
NM_001077262
NM_183008
|
UBXN11
|
UBX domain protein 11
|
|
chr16_+_69984809
|
0.448
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr2_+_177053306
|
0.446
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr10_-_131909061
|
0.446
|
|
LOC387723
|
uncharacterized LOC387723
|
|
chr8_-_103666018
|
0.446
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr9_-_35815040
|
0.443
|
NM_032593
|
HINT2
|
histidine triad nucleotide binding protein 2
|
|
chr17_-_79980685
|
0.441
|
NM_144998
|
STRA13
|
stimulated by retinoic acid 13 homolog (mouse)
|
|
chr19_-_10491212
|
0.439
|
NM_003331
|
TYK2
|
tyrosine kinase 2
|
|
chr19_+_55851220
|
0.438
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr4_-_174255454
|
0.438
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr19_-_4535202
|
0.437
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr3_-_141868208
|
0.437
|
NM_001178138
NM_001178139
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr7_+_129710572
|
0.436
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr5_+_64859098
|
0.435
|
NM_015342
|
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1
|
|
chr17_+_46018888
|
0.433
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr17_-_70116928
|
0.433
|
|
FLJ37644
|
uncharacterized LOC400618
|
|
chr1_+_150459913
|
0.427
|
NM_025150
|
TARS2
|
threonyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr19_+_17581276
|
0.424
|
NM_198580
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1
|
|
chr4_-_1722978
|
0.423
|
NM_001127266
NM_138385
|
TMEM129
|
transmembrane protein 129
|
|
chr19_-_14640048
|
0.423
|
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr7_+_99070428
|
0.423
|
|
ZNF789
|
zinc finger protein 789
|
|
chr6_-_32920812
|
0.421
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr8_+_118532970
|
0.421
|
|
MED30
|
mediator complex subunit 30
|
|
chr12_-_56211484
|
0.421
|
|
SARNP
|
SAP domain containing ribonucleoprotein
|
|
chr16_+_103859
|
0.421
|
|
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12)
|
|
chr12_+_50017340
|
0.420
|
NM_012272
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae)
|
|
chr22_+_19701968
|
0.420
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr15_-_83876165
|
0.420
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr16_-_2723374
|
0.416
|
|
ERVK13-1
|
endogenous retrovirus group K13, member 1
|
|
chr12_+_58087737
|
0.415
|
NM_001017956
NM_001017957
NM_001017958
NM_006812
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin
|
|
chr17_+_61627841
|
0.415
|
|
DCAF7
|
DDB1 and CUL4 associated factor 7
|
|
chr10_-_101380217
|
0.414
|
NM_031212
|
SLC25A28
|
solute carrier family 25, member 28
|
|
chr1_+_149824180
|
0.413
|
NM_021059
NM_001005464
|
HIST2H3C
HIST2H3A
|
histone cluster 2, H3c
histone cluster 2, H3a
|
|
chr20_-_32274154
|
0.413
|
NM_005225
|
E2F1
|
E2F transcription factor 1
|
|
chr19_-_14629160
|
0.412
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr16_-_30773303
|
0.412
|
NM_001014979
NM_001195620
|
C16orf93
|
chromosome 16 open reading frame 93
|
|
chr19_-_38720312
|
0.411
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr18_+_44526786
|
0.408
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr16_-_1876792
|
0.405
|
|
HAGH
|
hydroxyacylglutathione hydrolase
|
|
chr16_-_68482373
|
0.402
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr3_+_121554029
|
0.402
|
NM_018456
|
EAF2
|
ELL associated factor 2
|
|
chr21_-_47878592
|
0.401
|
|
|
|
|
chr16_+_838610
|
0.401
|
NM_022092
|
CHTF18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr19_-_36545646
|
0.398
|
NM_152658
|
THAP8
|
THAP domain containing 8
|